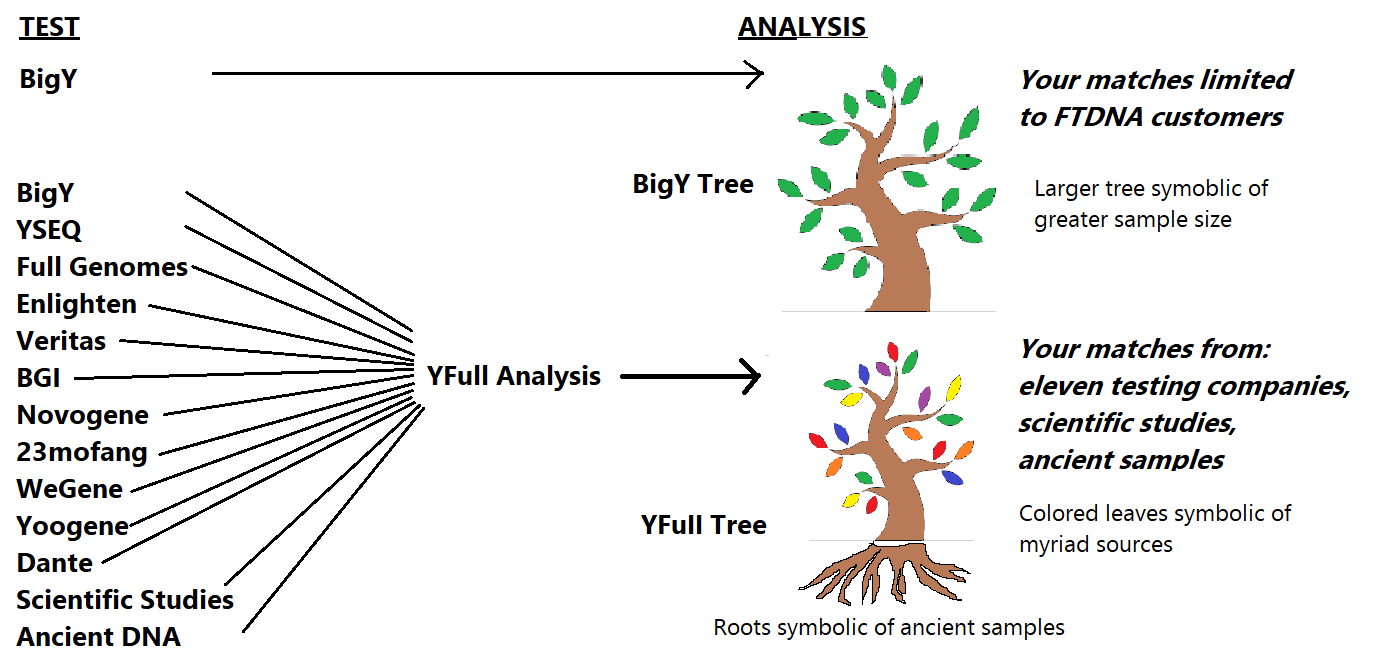
Big Y is a test. However, a test is meaningless without being compared to other samples.
FTDNA does an in-house analysis of your Big Y one view of which is called the Block Tree (available to customers) and the haplotree (public). You will only see FTDNA customers on this tree. YFull offers an analysis that compares your sample to those from eleven testing companies, including FTDNA, who have purchased the service for $49. The YFull analysis also provides a number of additional features outlined in the table below.
So while there are other deficiencies in the FTDNA Big Y Tree (see diagram below), the main reason Big Y customers do the YFull analysis is because it simply makes sense from a cost-benefit analysis to see relatives from all testing companies for just a fraction of the cost of the original test.
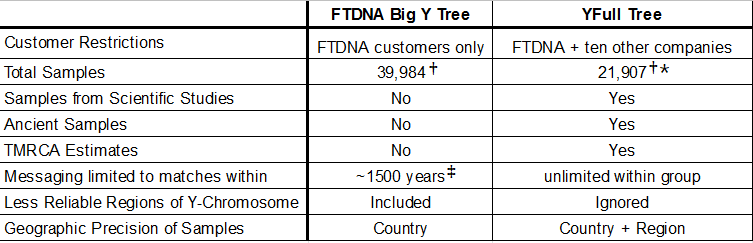
Customers have spent at least $500 on the Big Y and decided it was worth it to spend an additional $49 to get the best analysis possible - seeing relatives from all testing companies in a tree that also contains scientific samples and ancient samples.
An additional advantage is that any geolocated sample on the YFull tree contributes to public research into our common origins by means of the PhyloGeographer theoretical migration path project.
Going Forward - FTDNA Introduces $99 Tariff to do YFull Analysis
While FTDNA has maintained a good chunk of the Y-chromosome NGS market, competition is increasing. Dante Labs has attracted thousands of customers with its Black Friday sales, although anyone considering testing with them should know that they have failed to fulfill many orders in a timely fashion. Many of these samples are going on the YFull tree, none of them are going on the FTDNA Big Y Tree. With the rise of Nanopore technology there will inevitably be new competitive challenges.
To stay competitive against Dante Labs, FTDNA has announced it will slash test prices but tack on a $99 tariff for those who wish to do the YFull analysis. Of course it is ludicrous that this tariff, disguised as a "fee", is double the cost of the YFull analysis itself. This move may backfire as haplogroup researchers who always recommend the YFull analysis as the best value option for testers will need to take this additional tariff into account. The overall best value option may end up being Dante Labs if they get their act together and consistently fill orders and provide free BAM download link. If not, some other competitor.
The $99 Tariff is Bad for Haplogroup Research
Haplogroup researchers should understand that given this new $99 BAM download tariff, it will be more difficult to convince Big Y customers to do the YFull analysis. As a result, if the Big Y market share stays the same or increases, the pace of haplogroup research, in so far as it is defined by the YFull tree, will decrease.
The extent to which the YFull tree is perceived as the standard in haplogroup research varies from haplogroup to haplogroup. In J2 the consensus is that YFull is the standard - we value the additional samples from other testing companies and in particular the scientific studies in South Asia which make up 23 of the 38 total samples in J-Z2432.
Some haplogroup administrators have from the beginning not advocated their members to do the YFull analysis. It's never too late to recommend to your members to do the YFull analysis. But this is a daunting task for volunteer researchers of larger projects, so many may feel they can only continue research effectively from within the FTDNA data silo.
While their haplogroup research must continue within FTDNA unless/until they do they YFull analysis, these data siloed haplogroups need not completely endorse FTDNAs steps to monopolize the testing industry.
STR Match Finder is a free tool that allows people to find their STR matches within public FTDNA and YSEQ projects. While the matches displayed are limited to project members, the genetic distance cutoff is adjustable and the visualization helps easily identify potentially related individuals by showing rare matching STRs in shades of red. I used it recently to predict that a J-M241 Azorean would end up being the first European from otherwise South and West Asian J-Z2432 rather than belong to the J-L283 haplogroup more common in Europe.
While the FTDNA matching interface can show additional matches that have not joined a project, their genetic distance cutoffs are limiting for haplogroup researchers. I've found dozens of relationships in this way that would otherwise remain hidden within the FTDNA data silo.
FTDNA's $99 3rd Party Analysis Tariff Is a Two-Pronged Strategy To Monopolize the Testing Market by Undermining the YFull Tree
I perceive FTDNA's monopolization strategy as such:
- Less Big Y customers will do the YFull analysis given its steeper $148 effective cost
- The YFull tree will grow more slowly than it otherwise would
- This will help (but not guarantee) the FTDNA Big Y Tree to continue to outperform the YFull tree in the single metric where it objectively excels - total number of samples
- If the relative gap in sample size actually increases, FTDNA hopes that more future Y-DNA testers will decide that their FTDNA Big Y Tree is nearly good enough, equivalent or superior to the YFull tree
- Given a narrower perception of increased value, some additional customers will be drawn to test with FTDNA because of lower total test + analysis cost relative to perceived value
The stronger any company's monopoly, the higher the prices for customers and less future innovation.
Keep this in mind even if you are from one of the haplogroups which is very well represented on FTDNA and do not see much of a gain in doing the YFull analysis. The YFull analysis is and has been making your tests cheaper because FTDNA has to be competitive with people testing in other companies. In fact, FTDNA would not have implemented the Block Tree / Haplotree views or numerous other improvements without the competition offered by other testing companies, facilitated by the YFull analysis.
Instructions to do the YFull analysis - This may need updating depending on the new FTDNA workflow to get BAM file.
Genetic genealogist Linda Jonas discusses in more detail how to make use of YFull's capabilities in this article.
YFull gave me a Huge genealogical break through! FTDNA gave me a lot of possible leads, but nothing solid. YFull found I had a Y-Haplogroup that I shared with only 2 other people (and they had same surname as me – one with slight variation of spelling that I would have missed). That along with TMRCA and their paper trail found where my ancestor was in 1760 and with the other person, all the way back to 1584. YFull should be used by all genealogist!
i think so , yfull is far better in terms of haplogroups , i did a small test in FTDNA 37y and i’m very disappointed not even my haplogroup they predicted they just gave me the str’s value , i will not see any point to do testing if not the big y, so for me is thumbs down to FTDNA.,
I’m still a bit confused as to how one goes about getting their novel variants placed on a tree; any tree. From the FTDNA standpoint, it sounds like you are on your own until someone else does a BigY700 and matches you. And if you look at the criteria used by ISOGG, its a wonder anything makes it on a tree.
I was lucky in that my BigY700 finished up a few months before they imposed the BAM download tariff. Downloading that file was the first thing I did once my results became available. The way I see it, this raw data is your genetic legacy and what you actually paid for by doing a NGS test. The FTDNA genome browser is not too bad, but I prefer to use the Broad’s IGV to examine my BAM file. This gives you a much better graphical view of total reads, quality, length, etc as well as being able to set landmarks and SNP labels.
I picked my top five novel variants and followed Thomas Krahn’s validation protocol using the DNA of my paternal fourth cousin. This technically covered the “second person” requirement and using YSeq as a “second independent laboratory”. These variants showed up almost immediately in the YSeq catalog and now can be ordered by anyone. But when wil they eventually find their way onto a tree?
Hi Dwayne. Like you said, your private SNPs will show up on a tree – either FTDNA or YFull – once you have a match (the ones you share with another man/men).
Have you tried looking up your terminal subclade on YFull to see if there are additional samples there not present on FTDNA? My two matches are on the YFull tree and none of us are on the FTDNA tree.
https://phylogeographer.com/snp-lookup
Why don’t count the number per supplier/acadenmic paper that adds up to Yfull’s 21907 (at that time) test. Either they are extremely few as many states that yfull has half of the BAM’s from FTDNA, or it is few FTDNA tests in Yfull