Personal aside: My best friend from high school turned out to be R1a-L1029 several years back. His male line origin is quite different from the others, descending from a Syrian of assumed Albanian deeper ancestry based on the surname Arnaoot.
I was asked recently commissioned to do a new analysis of the geographic distribution of R1a-L1029 using HRAS by Hamit Koci.
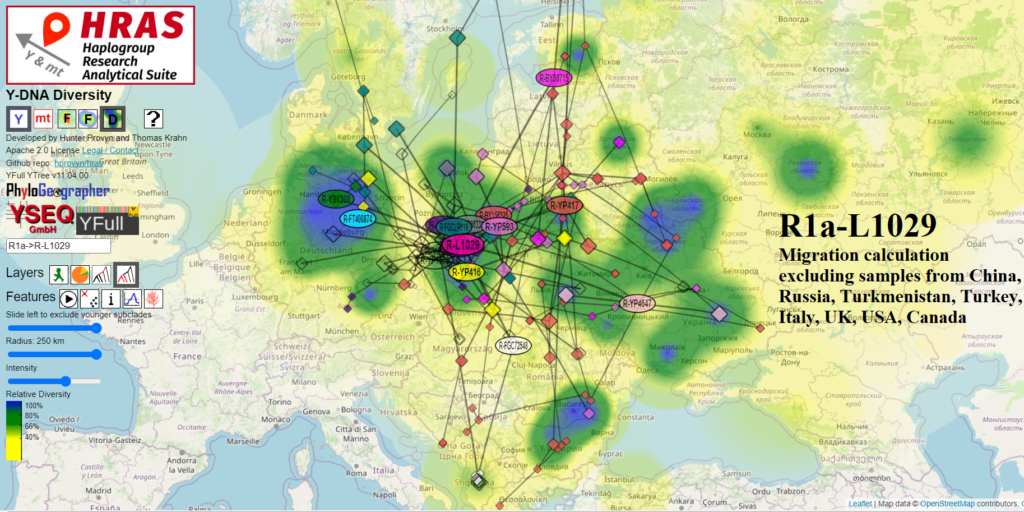
Iron Age Sample from Czechia implies 700-400 BCE TMRCA for R1a-L1029
I think that one very important development for R1a-L1029 research is the high resolution test of 2150-2350 years old ancient sample I13780 found in the Barrandov neighborhood of SW Prague.
YFull classifies this sample as R-YP5269, meaning that this ancient sample’s line had accumulated at least six SNPs after the R1a-L1029 TMRCA lived.
I didn’t try to learn about how this sample was dated, but if we consider the stated estimate of 2150-2350 ybp as more reliable than YFull’s estimates, this implies that R1a-L1029 itself may actually have a TMRCA several hundred years earlier than the 2100 ybp currently estimated by YFull. Given that YFull’s estimate for R1a-YP5269’s TMRCA as 1800 ybp is about 450 years younger than the ancient sample, if we tack these 450 years on to the 2100 ybp that YFull estimates for R1a-L1029, the adjusted TMRCA would be 550 BCE.
FTDNA currently estimates the TMRCA for R1a-YP5269 as 50 BCE and R1a-L1029 as 350 BCE. Given that the ancient sample is dated to 200 years older than FTDNA’s TMRCA estimate for the subclade, if we tack these 200 years on to R1a-L1029’s predicted age, the adjusted TMRCA would be 550 BCE.
Note that I’m not an expert on computing TMRCAs and that what I did was a simplistic back of the envelope computation. However, I think it is interesting that using the same method to adjust the TMRCAs results in the same date of 550 BCE. I arbitrarily added a +/- 150 year error interval to this figure in the heading above.
La Tene Bohemia is geographically peripheral to the center of R1a-L1029 expansion
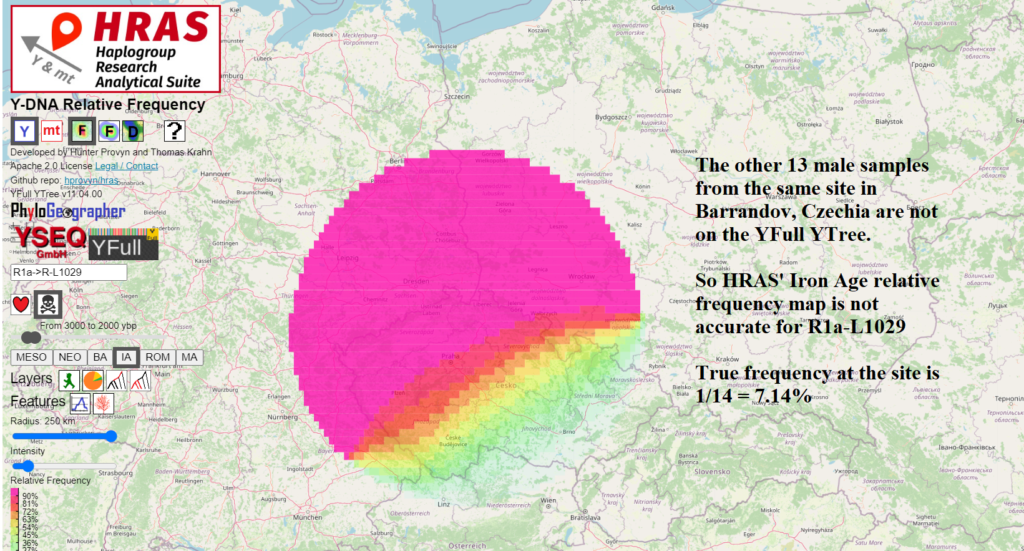
Of the 14 samples from the site in Barrandov, Czechia with male lines entered on the Ancient Human DNA Map, only one is R1a-L1029. So from this site, we have a relative frequency of 7.14%.
I share the Iron Age relative frequency of R1a-L1029 computed from HRAS below simply to show that this functionality exists – however the result is inaccurate in this case because only 1 of the 14 male line samples from the Barrandov site is on the YFull YTree, the data source for HRAS. However, you may wish to use the HRAS ancient relative frequency functionality for other haplogroups with better ancient representation on the YFull YTree.

While the area of Prague appears to be in or near one of the three core La Tene centers from the map from Wikipedia, this location is peripheral to where HRAS estimates R1a-L1029 and most of its subclades to have formed, in and around the Łódź Voivodeship.
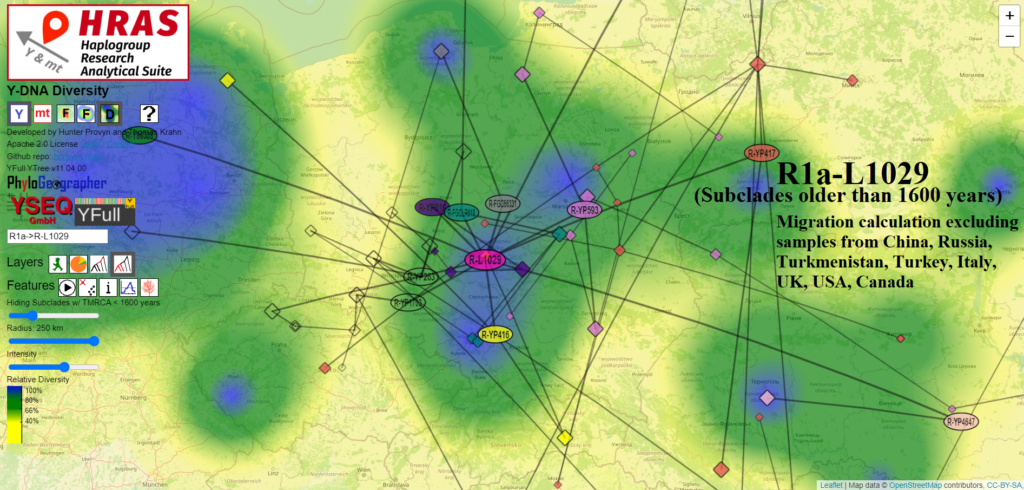
Given that R1a-L1029 may have diversified by 550 BCE in or around Łódź Voivodeship, it would make sense to view this lineage as one that began diversifying prior to the arrival of Celtic influence in its southwestern periphery several hundred years later.
R1a-L1029 and the ethnogenesis and migration of Slavs
HRAS also features a diversification-over-time chart. The diversification-over-time chart for R1a-L1029 shows that the growth of these lineages was greatest in 550 BCE (or whenever the actual TMRCA lived) and that growth continuously decreased ever since then, with no local maxima of increased growth.
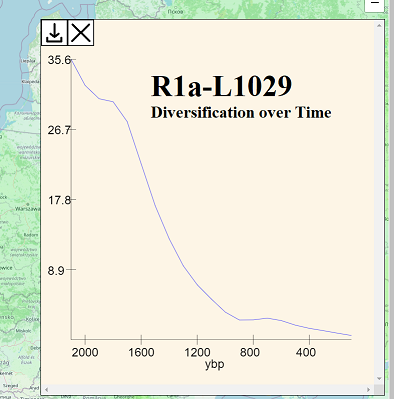
If other lineages that are also considered as having been core to the ethnogenesis of western or eastern Slavs show increased diversification relative to R1a-L1029 during the first several centuries CE, we might draw the conclusion that R1a-L1029 had lower fitness due to being politically subjugated by other Slavic/Proto-Slavic coalitions richer in other haplogroups.
Prolific R1a-YP417 estimated to have originated in NW Zhitomir, Ukraine
This lineage is more prevalent in Russia and less prevalent in Poland than other lineages of R1a-L1029.
| Poland | Russia | |
| R1a-L1029(xYP417) | 66 | 25 |
| R1a-YP417 | 5 | 63 |
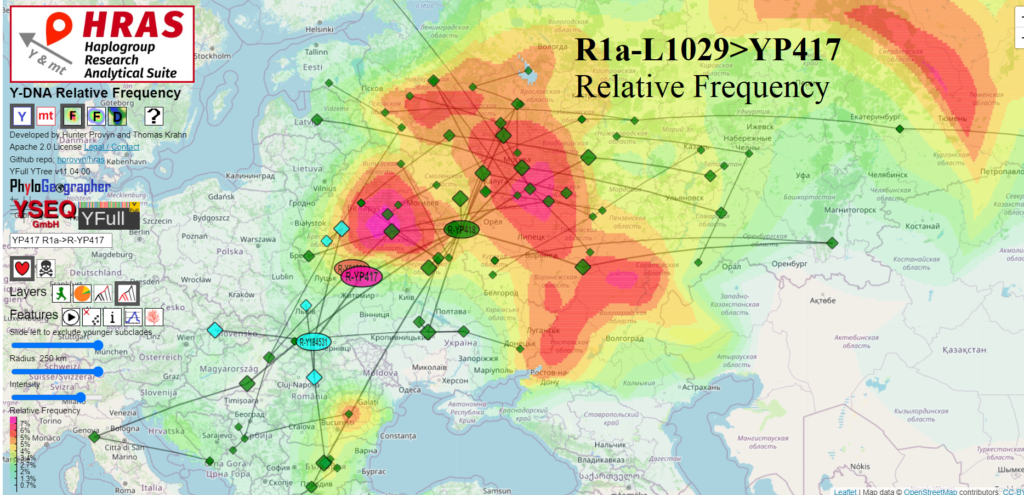
Actually only one of the three subclades of R1a-YP417 is found in Russia, R1a-YP418. And interestingly the only samples from Poland on the YFull YTree so far in R1a-YP417 are in the same prolific child R1a-YP418. The geography that the other two branches of R1a-YP417 have in common is Belarus and Ukraine. That is why HRAS computes the origin of R1a-YP417 to NW Zhitomir Oblast, Ukraine, without setting any countries’ samples to be excluded from the computation.
However, because only three total subclades exist, one of them with a TMRCA of only 450 ybp, this computed origin is less reliable than the R1a-L1029 computed origin with 16 subclades / basal samples.
This line appears to have been under some pressure to migrate east or perhaps it was offered to rule a newly conquered region in this direction, starting with the R1a-YP417 TMRCA who may have been born after the migration of his parents to NW Ukraine or traveled there himself as a child from the R1a-L1029 potential approximate homeland around Łódź, Poland.
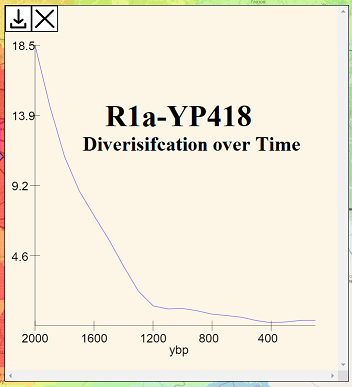
R1a-YP418 – Sire of 12 lineages – possibly Zarubintsy culture
R1a-YP418 sired at least 12 lineages. Almost as many as his ancestor R1a-L1029, founder of this dynasty, who sired at least 16. I do not believe that periods of relative demographic growth alone can explain such outliers in terms of fecundity. I believe these men wielded considerable political / military power AND may have also been (but not necessarily) living in a time and place where the environment was conducive to growth.
Given the large number of lineages sired by R1a-YP418 and geographic consistency further to the east, it makes sense that this lineage held considerable power around the approximate area of Bryansk, Russia only 2 SNPs after the R1a-L1029 founder lived. He would have lived around 250 BCE if we round 144 years per SNP to 150 and add 300 years to the 550 BCE figure I had computed above for R1a-L1029’s TMRCA. Turning this into an interval it could be 400-100 BCE.
The only culture I could find in the approximate area between Ukraine and Bryansk for this period is the Zarubinsty culture which existed from 3rd century BCE to the 1st century CE according to Wikipedia. This culture was influence by the La Tene culture of the Celts and the steppe nomads (Scythians and Sarmatians). After the decline of this culture the inhabitants were incorporated into the Kyiv culture and Wielbark culture according to the article on Wikipedia.

R1a-L1029>BY30007>FGC72548>Y133361 – Estonian and Gheg (northern) Albanians
Note that on the FTDNA tree, BY30007 defines a branching point directly below R-L1029 and including children YP263 and FGC72548. YFull does not use this SNP for their tree.
The scarcity of samples that have set a paternal ancestor origin in R1a-L1029>FGC72548 on YFull offers no clues to the deeper origin. Both Estonia and Albania are extreme outliers to the core R1a-L1029 diversity.
I will note that there are two FTDNA Big Y samples at the position of R1a-FGC72548*, one tracing to Germany and another to Czechia. The German has no STR matches in public projects and the sample from Czechia is unknown because he didn’t join any public projects.
The good news for our research is that the Albanians in this line are very distantly related to one another. YFull estimates that their MRCA lived around 1200 years ago.
Since the two lines diverged dramatically around 1450 years ago, or 570 CE, we can be reasonably certain that their MRCA was still living somewhere around the core R1a-L1029 area at least until 570 CE.
The migration of the Albanian line R1a-Y133367 could have taken place any time between 570 – 820 CE, if it was the migration of a single man who only diversified once in Albania.
So I think it makes sense that the ancestor who migrated to Albania did so during one of the waves of Slavic raids or migrations during that period. This/these ancestors may have co-migrated to Albania together with a branch of R1a-L1029>YP263…
R1a-L1029>BY30007>YP263
This lineage has a much more well defined substructure than its sibling, due to having had many more surviving descendants.
I share two screenshots from HRAS depicting the approximate theoretical origin of R1a-YP263: one including samples from Albania and the other excluding them. There is not a big different
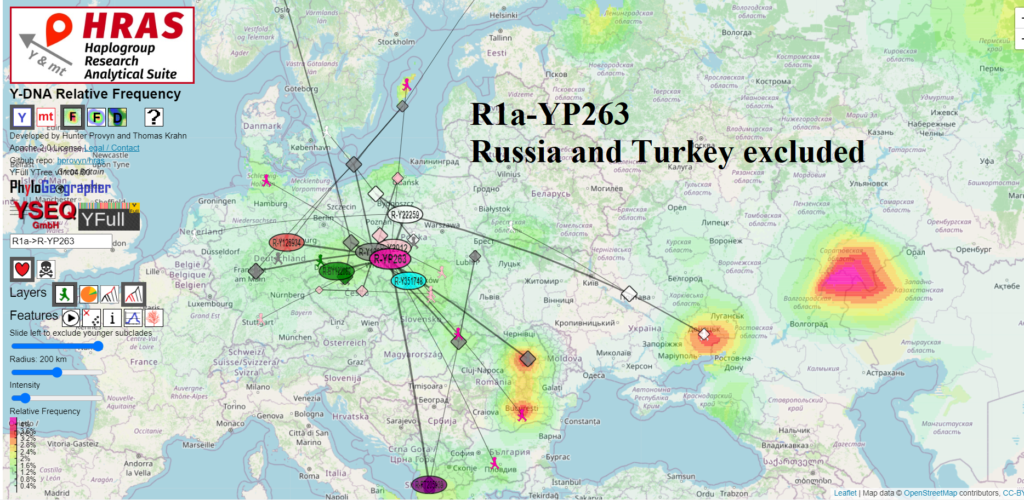
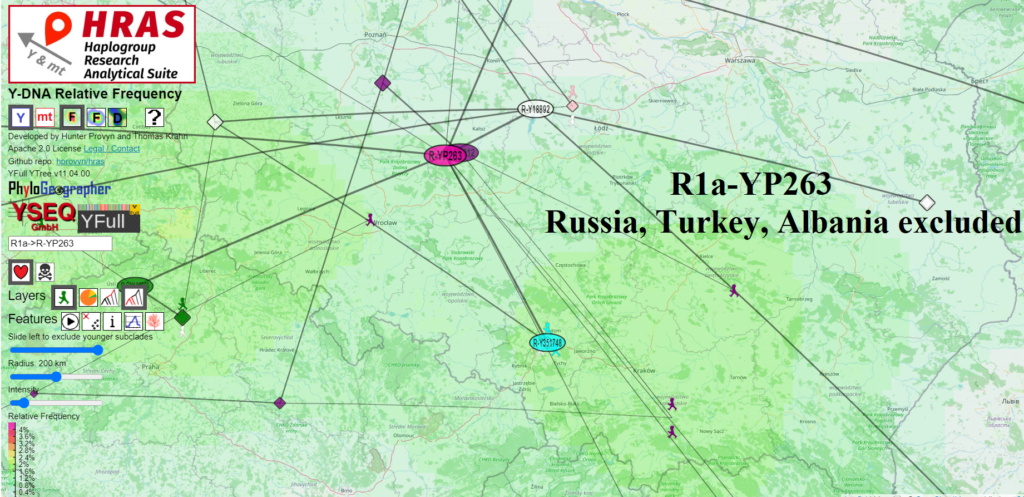
Excluding the samples from Albania, HRAS computes R1a-YP263 to have originated in the approximate area midway between Łódź and Wrocław. Including the samples from Albania shifts the center south about 40-50 km. This difference is nearly imperceptible unless you zoom in.
So R1a-YP263 does not appear to have originated significantly further south than R1a-L1029 (Łódź). However the picture may change depending on how the two new samples on YFull from Romania and Bulgaria affect the tree, both currently are are at the R1a-YP263* level.
So it may have been that one descendant of R1a-YP263 and one descendant of R1a-FGC72548 happened to have migrated somewhere further south than each of their closer relatives, possibly to the same general area, and from there these distant cousins became incorporated into the same / different groups that migrated to Albania. If the migration happened 1200-1350 years ago, this would have been about 1200 years after their MRCA R1a-BY30007 would have lived (having added 200 years to FTDNA’s estimate of 250 BCE).
STR Analysis of R1a-L1029 From Studies
In this section I use STR Match Finder which I developed to view rare alleles shared by relatively closer GD matches.
Unfortunately, the very low level of STRs tested by these studies usually would leave us with uncertainty as to the exact haplogroup of R1a-L1029 of most samples.
However in this case we are lucky, because the aforementioned Albanian line R1a-Y133367 have two rare alleles defining their line within the first 37 STRs usually tested, DYS389i (in the first 12 STRs) and DYS460.
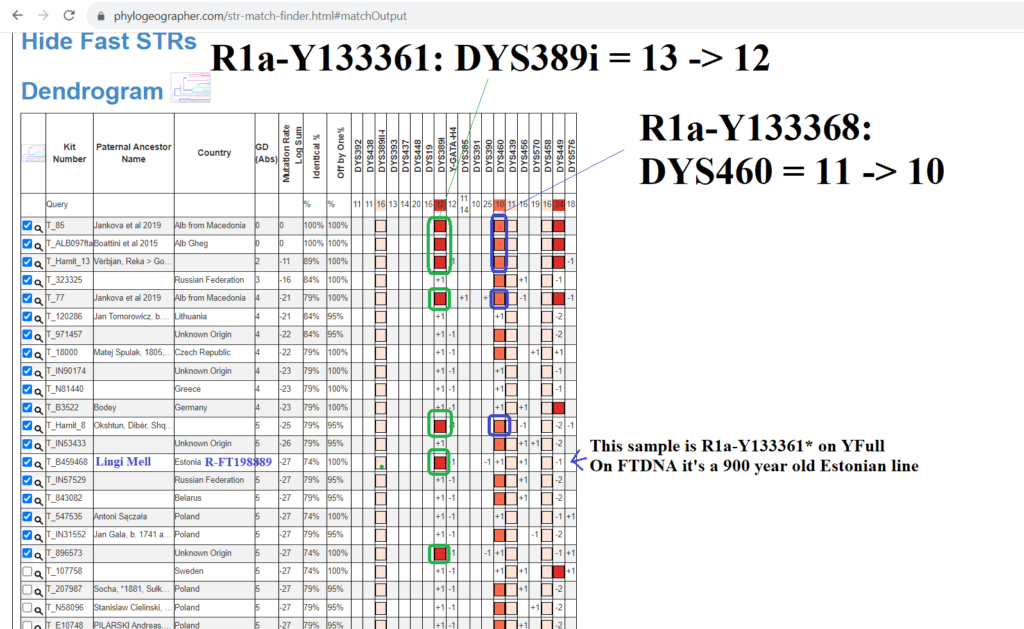
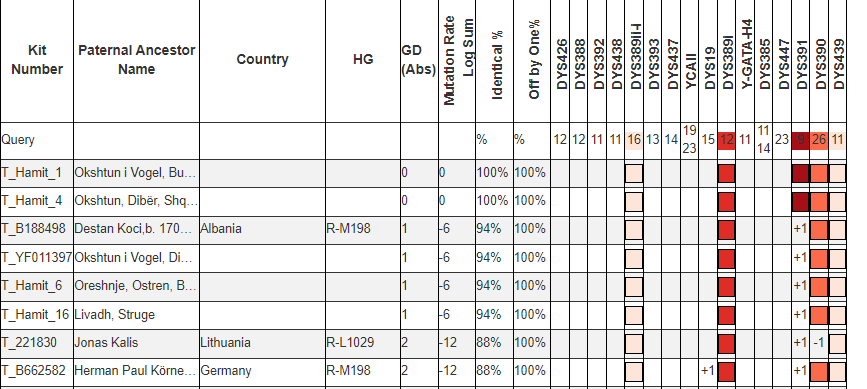
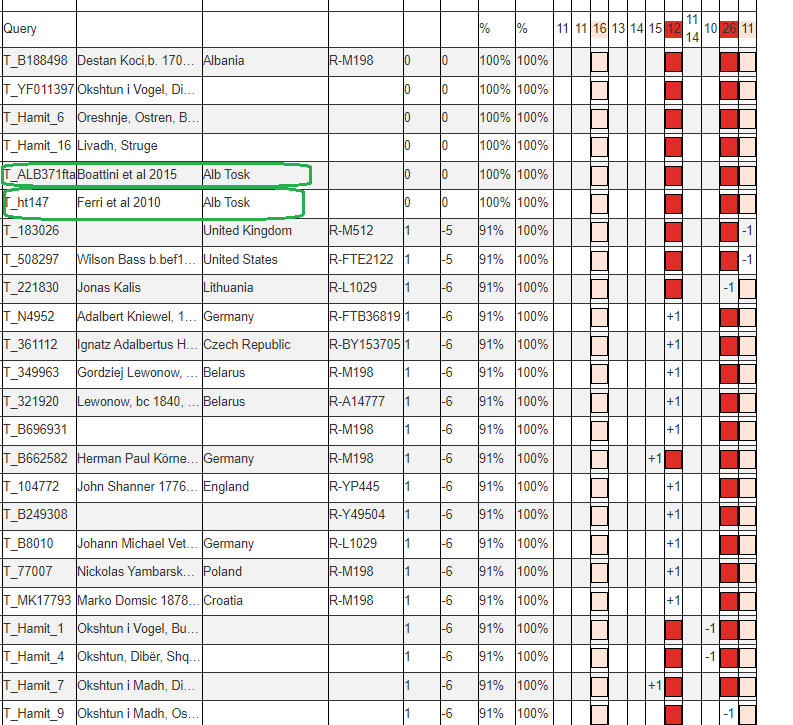
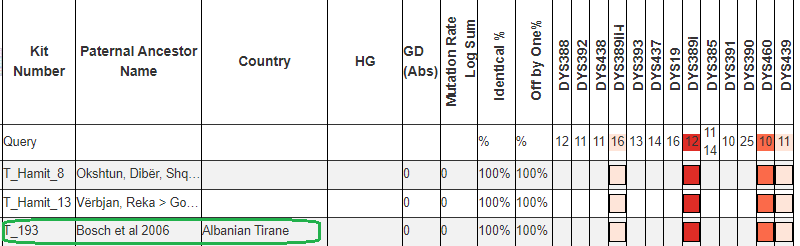
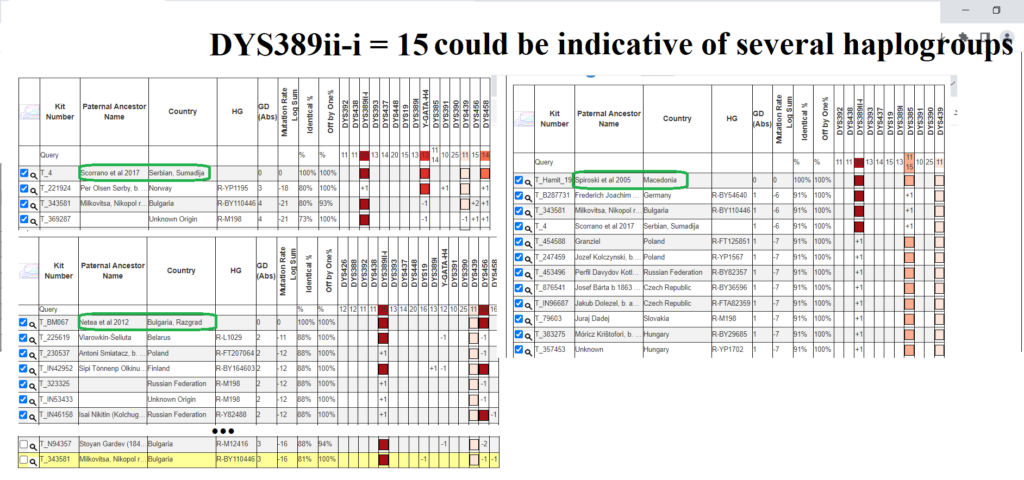
Matching a rare DYS389ii allele with another sample is not always indicative of more recent common descent. This is because DYS389ii is the sum of two different alleles that can mutate independently, DYS389i and DYS389ii-i, for lack of a better name. While these men have DYS389ii = 28 like the men in R1a-Y133361, they are likely not actually related to them because they didn’t arrive at DYS389ii = 28 from having DYS389i = 13 -> 12 mutation. Instead their DYS389ii-i allele decreased. Both independent mutations result in the same sum for DYS389ii.
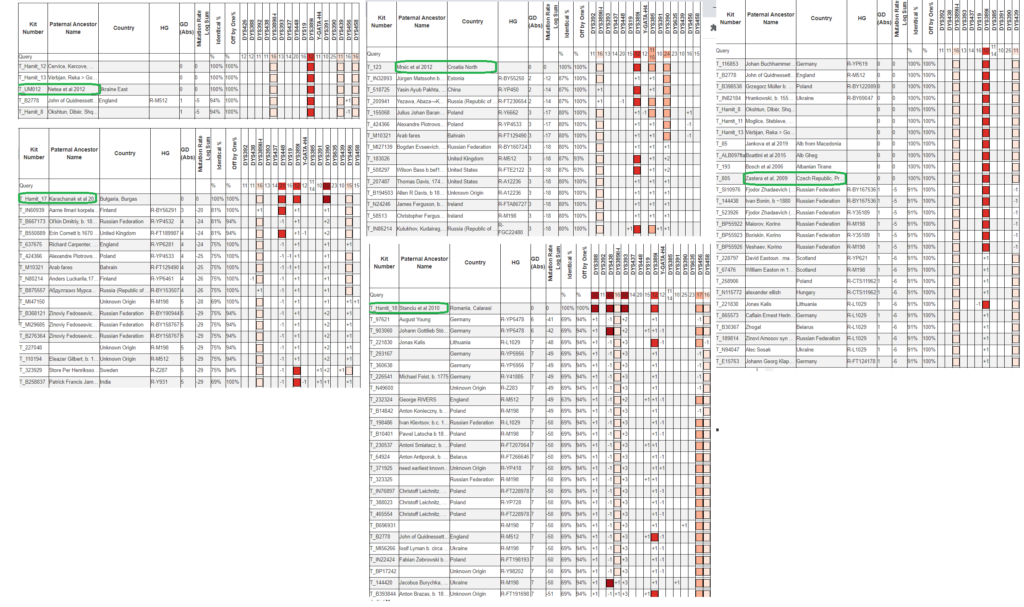
In the above screenshot of STR Match Finder screenshots, in the top left we have results for UM012 from Netea et al (2012) from East Ukraine. While the only exact matches are Albanians in R-Y133367, there is also a match only off by one to a man of undetermined haplogroup likely below R1a-L1029, B2778 of England. This sample has tested many STRs, some from the 111 but not all for some reason, yet I cannot predict which branch of R1a-L1029 he may be. I’d recommend testing him for a SNP at the level of R-Y133361 or higher (R-FGC72548) in case he ends up splitting R-Y133361 (i.e. being positive for some but not all the SNPs at that level). Back to UM012, he may indeed be more closely related to R-Y133367 but it’s not for certain given the extremely low level of STRs tested.
On the top right are the matches of sample 805 from Zastera et al (2009). Because he has tested only eleven STRs he has exact matches among men in different haplogroups of R1a-L1029. Therefore he could be more closely related to any of these men and even to men that he doesn’t exactly match in this small set of eleven.
The other men do not appear more closely related to R1a-Y133367 by STRs and might be related to some of the men among their closer STR matches as indicated.
These posts are the opinion of Hunter Provyn, a haplogroup researcher in J-M241 and J-M102.
thank you for this study
Thank you for this research – very interesting!
I’m from Estonia, R-Y133361* / R-FT198889
You are welcome, Alexander. I’m glad you found the maps and analysis interesting.
Hi,Im Milan Sefcik from Czech Republik.
I have Y700 FF, haplogrup YP445.
Note that settlements of the La Tène culture came as far as Serbia, Bulgaria and even to ancient Anatolia (Galatia) and also areas like Silesia (todays Poland) and there have been extends of La Tène influences found as far as the Black Sea. Balkan Celts is an interesting blog to see were this culture and it’s people moved around (an irish guy who is in Bulgaria I think). The trade connections to the Baltic for Amber were quite long lasting as well, but don’t seems a point of origin to me, from my research. Btw. most who are close to my Haplogroup are overwhelmingly from Germany and Germans had a history of settling all these areas of L1029 as well. Just my 2 cents.
I wondee how do YFull and FTDNA fit all ancient samples into the databese that is used to calculate SNP ages. Do they have a special provision that lets yhe computer understand those are ancien samples? Or do they not use them at all? Or do they enter them as recent living people? If the latter, then ofcourse the calculations would be flawed.