In 2018, Di Cristofaro and others published Prehistoric migrations through the Mediterranean basin shaped Corsican Y-chromosome diversity. In their table they provide a limited set of STRs from Corsican samples that they collected alongside samples from nearby regions from other studies.
One thing distinguishing the samples of King (2011) and Di Cristofaro (2018) from Boattini (2013) was that they were tested for DYS445 and DYS461. Because a rare DYS445 allele is associated with one basal and one predicted early/basal J2b-L283 sample I restricted the comparative STR analysis to those samples with a read.
The set of STRs tested for these samples: DYS456, DYS389I, DYS389II, DYS390, DYS458, DYS19, DYS385, DYS393, DYS391, DYS439, DYS635, DYS392, YGATAH4, DYS437, DYS438, DYS448, DYS445, DYS388, DYS461
Counting palindrome DYS385 as one STR, it's a total of nineteen STRs.
Of the thirteen samples that have reads for all alleles, four are from Provence and nine are from Corsica. The latitude and longitude for all Provence samples corresponds to Marseille. Six of these samples were classified as J2b-M12 but appear to be J2b-L283. There was no note indicating they were confirmed M241 negative, so I assume they are in fact J2b-L283.
To get an idea of how the samples are related to one another, I applied scipy's hierarchical clustering using a precomputed distance matrix. The distance matrix I supplied was the mutation rate-weighted log sum of STR differences from one sample to another as computed by STR Match Finder.
I included additional samples representing the major branches of J2b-L283 in order to see how the samples from the study cluster with them.
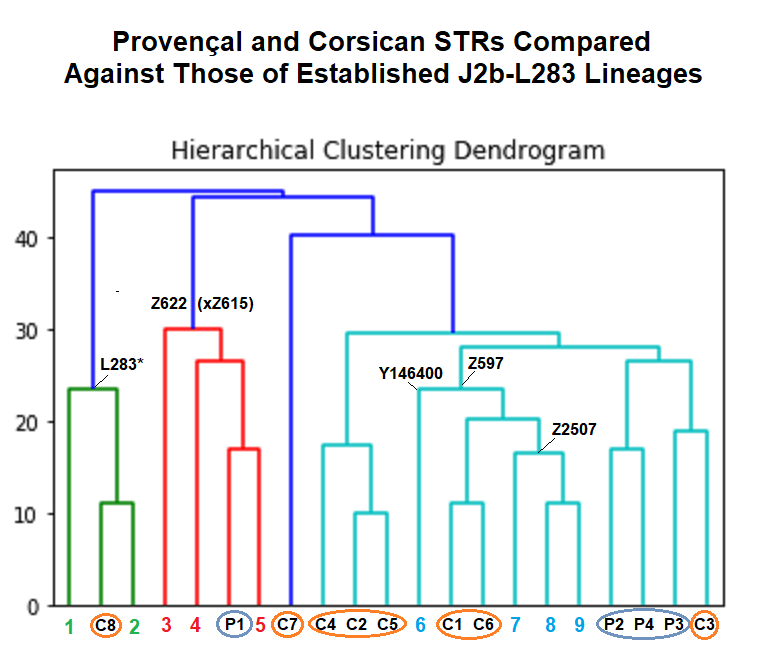
I am actually shocked that at such a low number of STRs compared, the clustering algorithm organized the samples into a tree that correctly separated the SNP-tested samples into three major sections of the J2b-L283 tree: J2b-L283*, J2b-L283-Z622(xZ615) and J2b-L283>Z622>..>Z615>Z597.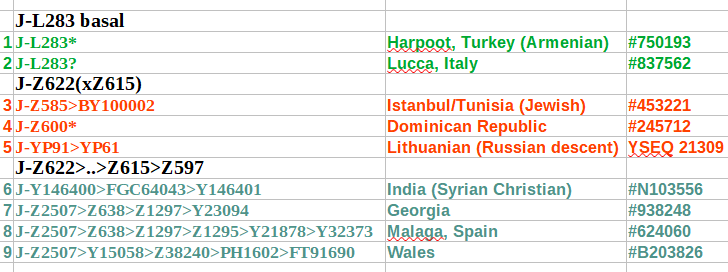
I think it helped that two of the samples from the study, C8 and P1, have the off-modal allele DYS437 = 15 which is ancestral for most haplogroups below J2b-L283 that do not descend from J-Z585.
C8, the sample collected in the study from CAP, Corsica, has high affinity to the predicted early/basal J2b-L283 from Lucca, Italy. They differ on just two STRs out of the 19 (DYS635, DYS439) and the man from Lucca is this Corsican's closest mutation rate weighted genetic distance match out of all samples I know of, because these alleles are higher variability.
Their geographic proximity is an encouraging sign that they probably represent the same early or basal lineage of J2b-L283.
P1 from the King study from Provence with DYS437 = 15 is another interesting sample. Based on his high divergence to other samples and this allele, he may be somewhere above J2b-Z585, though whether he really is J2b-YP91 like his closest match in the dendogram is less certain. Interestingly I notice that most of the Poles who are predicted J2b-YP91>YP61* also share his DYS19 = 16, so if they were to test the other alleles it may show a higher affinity to them.
The most divergent sample of this group on the dendogram is C7 from BASTIA. However given the small number of samples used to construct the dendogram and the small number of STRs, this is not strong evidence that he would actually be Z597 negative, but if we had access to his sample (we don't he's anonymous from the study) I would nonetheless invest in testing his SNPs to check.
C1 and C6 likely are each other's closest relatives, sharing rare DYS461 = 11 and the only two samples from TRAVU. The sample from the project that is J2b-PH1602 that I added is one of their closest matches in STR match finder and shares their off-modal DYS456 = 12 that most J2b-PH1602 have. However the result of the clustering grouped this Welsh man more closely together with the two men who were J2b-Z2507>Z638 rather than with these Corsicans. He also grouped closer to these men than to the other two samples with DYS456 = 12, C2 and C5, closely related and the only from AJACCIO. Some of these men could still actually be J2b-PH1602, I only added this one man to compare against them in the dendogram.
I'm not sure how significant the result is that there are two exclusively Corsican/Provencal out groups that cluster with each other rather than with any of the samples from our research project who are positive for various lineages below J-Z597. In each case I picked the samples from our project based on their showing up as proximal matches to Corsicans and Provencals on STR Match Finder, not for them being mutual matches.
Also that all three samples from Provence with DYS437 = 15 form a cluster with one Corsican on the far right may not be very significant given that this cluster's height (a measure of how distant the samples from the two children lineages were from one another when the branches merged) is almost as high as the next two levels that join the bulk of the samples with one another.
Because none of these samples in the teal group contain DYS437 = 15 they probably all represent lineages below J2b-Z585.
Going Forward
We plan to upgrade the man from Lucca to WGS / Big Y if he consents. Understanding his place on the YFull tree of J2b-L283 will give us a little more to work with for our theories.
Based on the two promising basal/early J2b-L283 samples it seems likely that Provence and Corsica contain samples that will advance the research into our common origins, if we are able to identify and advanced test the approximately 1/4 and 1/8, respectively, of J2b-L283 men who live there that are basal/early J2b-L283.
Not included in my STR analysis because he had no reads for DYS445 or DYS461 is Boattini 2013 sample 108. He is the only one from the study who was J2b-L283 and from Como. He appears likely to be negative for Z585 because he has off-modal DYS437 = 15.
So Como may be a promising place to collect pre-J2b-Z585 samples.