Check it out yourself at hras.yseq.net!
15 New Regional Codes
BD-20 Bangladesh (Habiganj) R-Y879
BR-RR Brazil (Roraima) J-BY127242
DO-01 Dominican Republic (Distrito Nacional (Santo Domingo)) E-Y224146
DO-22 Dominican Republic (San Juan) J-FTC45945
DZ-14 Algeria (Tiaret) E-Y218033
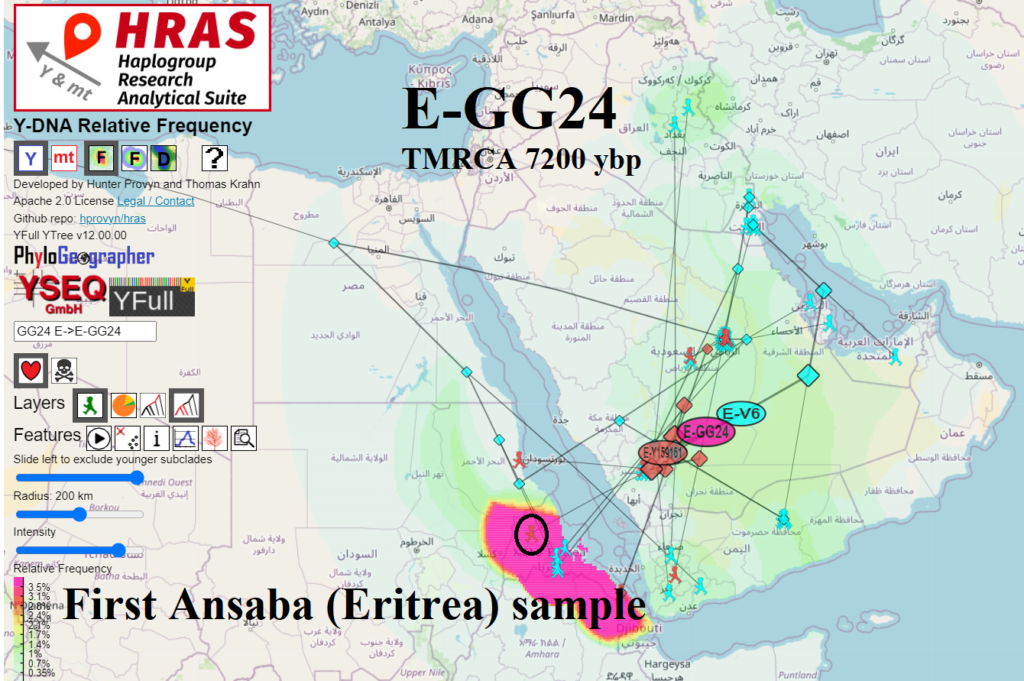
ER-AN Eritrea (Ansabā) E-FT373026
ER-GB Eritrea (Qāsh-Barkah) J-FGC83829
GB-WLV United Kingdom (Wolverhampton) R-Y302902
HU-HV Hungary (Hódmezővásárhely) R-Y317566
LV-074 Latvia (Priekules novads) R-S26229
MA-HAJ Morocco (El Hajeb) E-Y246379
TN-43 Tunisia (Sidi Bouzid) J-FGC38800
VC-01 Saint Vincent and the Grenadines (Charlotte) E-CTS2754
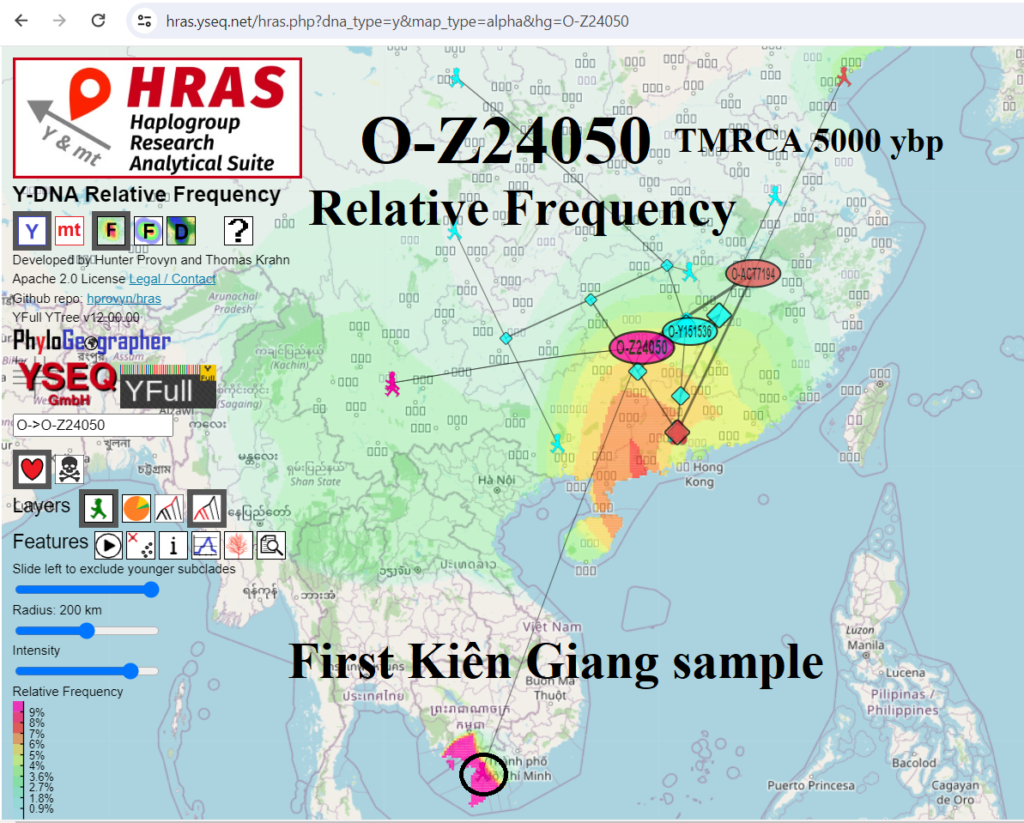
VN-47 Vietnam (Kiên Giang) O-Z24050
VN-48 Vietnam (Can Tho) O-V3237
Possible Next Features / Improvements
- Show TMRCAs of subclade and downstream node(s) on the map
- Improve migration path calculation algorithm:
- For outlier sample/nodes to relative to initially computed parent origin pathology* (this is most evident for a subclade with only two nodes)
- Take regional sample rate into account for weighting samples/nodes

*PhyloGeographer (forerunner to HRAS) has additional logic that, once subclade origins are computed solely on the basis of downstream sample/node locations (recursively, i.e. from bottom to top, starting from the nodes and moving up to the parents and grandparents, etc.), there is a second “refinement” pass whereby the initially computed subclades are used to determine which of downstream samples/nodes should be completely disregarded as outliers.
For the initial release of HRAS I left out the second step to start out with a simpler to code/understand/explain algorithm design, to ensure computation in the browser was not too slow, and to ensure that small changes to self-reported sample origins does not cause big changes to the computed origins.
I now feel confident to proceed with a 2nd refinement pass, using some of the newer methodologies I’ve developed since HRAS release to treat outliers with weights generated by a continuous function rather than the PhyloGeographer all-or-nothing (outlier = 0, non-outlier = 1) way, for which small changes to self-reported sample origins can cause big changes to computed origins.