I recently noticed that part of the code that was supposed to treat ancient samples differently had not been working correctly, due to a changed variable name during the refactoring process.
After fixing this, I realized that my intended method for assigning additional weight to ancient samples suffered from two flaws, both relating to the rule that small changes to input data should not result in major differences in computed output.
According to my original design, “ancient samples”, regardless of how old, would always have received triple the weight of their age-to-parent-TMRCA ratio submetric (itself a number scaled from 1-2). The problem is that an ancient sample that is only slightly older than a modern sample would have been counted three times as much as a modern sample, when we would actually prefer the weights to converge.
The other issue was that I didn’t add a mechanism for granting some additional weight to a child branch that is supported by ancient samples downstream of it. This caused a discontinuity where, if an ancient sample that originally was basal is later found to form a subclade, its weight would have shrunk to one third because branches were never given a bonus for containing ancient samples.
The new method I have implemented for assigning weights to nodes preserves the desired continuity.
The weight of a child node is now calculated as the sum of two submetrics, one relating to its TMRCA relative to the parent, the other relating to its oldest ancient sample age relative to the parent’s TMRCA.
These scores, as before, originally derive from a number from 0-1 (a ratio) that I translate to the scale of 1-2 (so that basal samples, treated as nodes w/ TMRCA 0 ypb, will not be worth zero). Another innovation I have introduced at this time is to square these submetrics before summing them together. This results in a weight on the range of 2-8 and one where relative age differences of younger branches / ancient samples do not affect the overall weight as much.
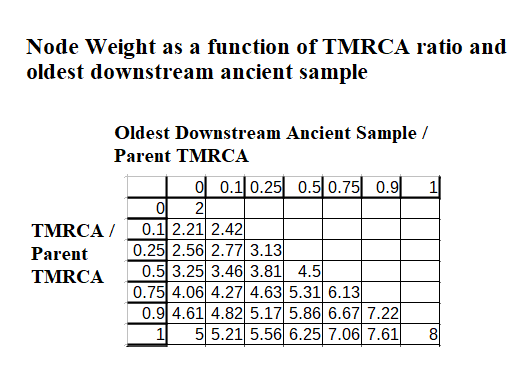
At one end of the scale, a basal modern sample has weight 2. At the other end of the scale, a basal ancient sample / branch containing an ancient sample, where the ancient sample is as old as the parent’s TMRCA has a weight of 8. So the most any one node (branch/basal sample) can be weighted relative to another is 4 times as much.
These posts are the opinion of Hunter Provyn, a haplogroup researcher in J-M241 and J-M102.