An alternative title to this post, using more technical language would be:
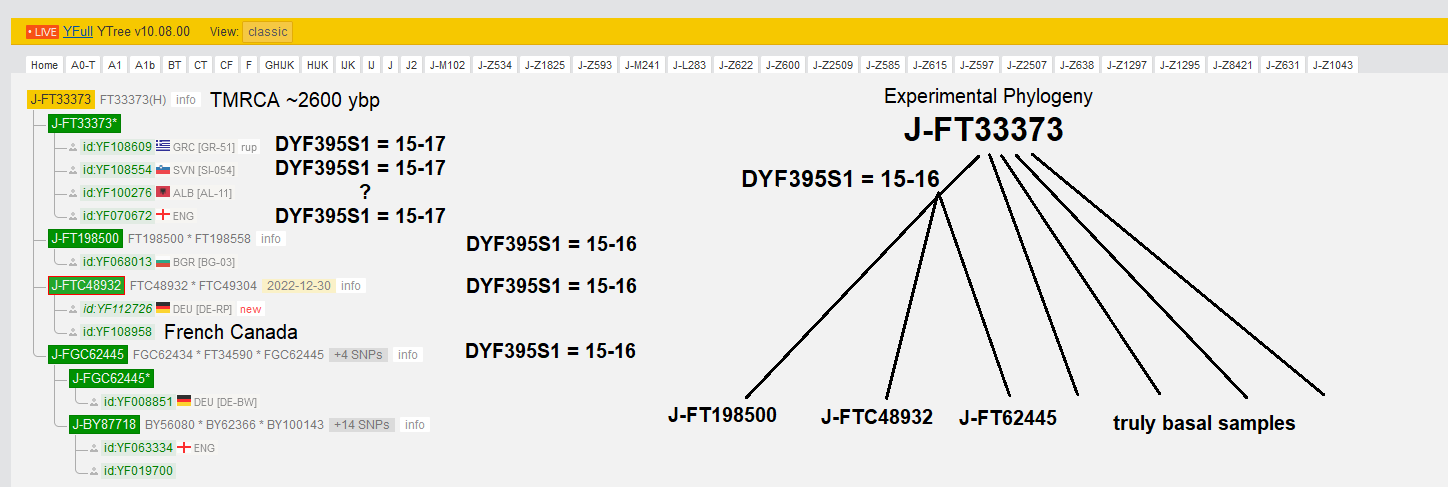
"Three subclades of J2b-FT33373 may actually descend from a single more recent common ancestor subclade of the parent"
A reliable SNP has not yet been found to indicate that these three child lineages all descend from a single common child line of J2b-FT33373.
That is why on the YFull tree, the three branches are considered siblings to each other and all the other basal J2b-FT33373 samples.
However the STR allele DYS395S1 likely did not coincidentally mutate from ancestral 15-17 to 15-16 independently in each of the three branches. Most men in J2b-L283 have DYS395S1 = 15-17, and more importantly the immediate parent J2b-Z1043 had 15-17.
Therefore I propose the above-pictured experimental phylogeny for J2b-FT33373, that might eventually be validated by higher coverage SNP tests of at least one man from two of the three child lines.
Especially strong circumstantial evidence of an early mutation to 15-16 is that both samples of J2b-FTC48932, which only share two SNPs with each other below J2b-FT33373, have this rare allele.
So the simplest explanation for this phenomenon is that these three lineages, established by SNPs, in fact descend from one subclade, for now only identifiable by DYF395S1 = 15-16, until a SNP can be found that they share.
I would recommend doing WGS for one representative each for two of these three lineages, J2b-FT198500, J2b-FTC48932, and J2b-FGC62445. So far, all of the men yet tested in these subclades have only done Big Y which systematically misses some parts of the Y covered by WGS tests, like this one at YSEQ that I recommend.
Conclusion
The impact of this discovery, if it turns out to be true, is that the non-Balkan diversity of J2b-FT33373 decreases slightly.
Two non-Balkan subclades and one Balkan subclade collapse into a single clade that is now weighted as much as any of its siblings - the basal samples, three of four of which are from the Balkans.
Thank You
We were able to upgrade the Vlach male line sample from Grevena, Greece and the line from Krško, Slovenia to WGS thanks to donations from the J2b-L283 research community.
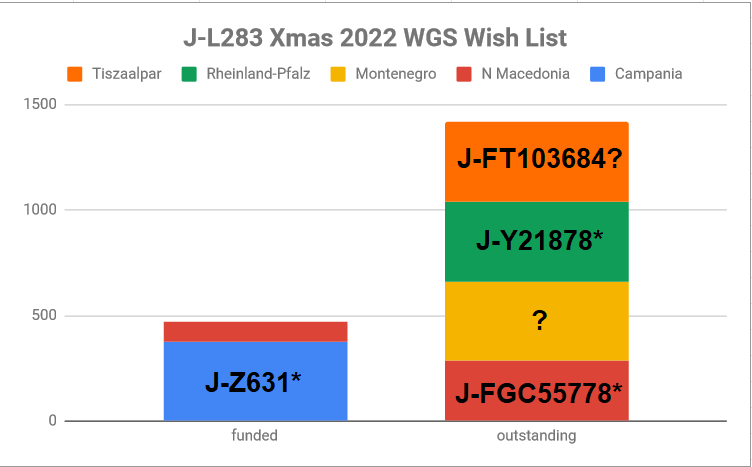
I have an ambitious plate of candidates I'd like to upgrade now. Two of the five I have already purchased WGS for and I wait for donations to cover any of the remaining to proceed further.
Sorry, off-topic question, but do you know how accurate are the TMRCA dates for mtDNA on YFull? Mine says 300ypb, which seems awfully recent so I’m not sure how accurate it is. Also, what’s the difference between formed and TMRCA? How do they know when the clade formed? https://www.yfull.com/mtree/U4a2c2a/
You can ask in the YFull group on FB, but I believe the mtDNA TMRCA estimates are more of a work in progress and less reliable than the Y-DNA TMRCA.
If you go up the tree you’ll notice that the formed estimates are identical to the TMRCA estimates of the clade’s parent. Formed means when this subclade began branching off from the parent, i.e. when the mutations that define it began to accumulate.