10/21/2020
I've made some major improvements to STR Match Finder.
I've added an instructional video.
Older
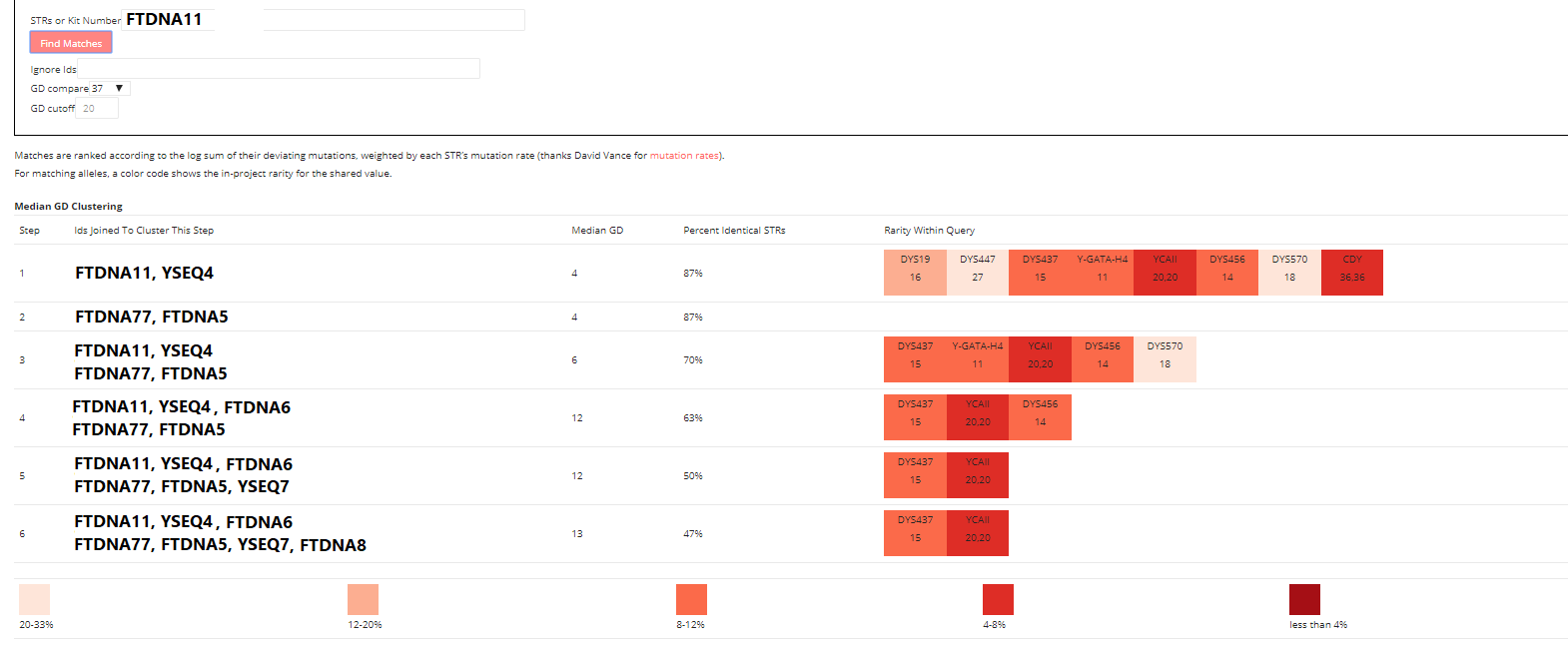
I've added automatic group discovery, technically a form of median clustering, to the STR Match Finder.
This is a tool you can use to find your relatives within public FTDNA and YSEQ projects.
The new feature groups samples with samples [and then groups with groups], at each step minimizing the median pairwise genetic distance of the resulting cluster.
I will probably try to add some tree-type graphic visualization and also incorporate country info. It's still in development and I need to add some settings to make it more robust.
The attached screenshot is a highly divergent Polish/Ukrainian cluster within J-L283>YP91 that I had recently identified by visual inspection of the STR Match Finder output. The IDs have been changed to fake IDs.
As a test, these men, though extremely divergent at absolute GD 12/37 STRs, do end up clustering together as expected, given their high number of shared rare STRs. Note that none of these men have yet to do NGS testing to prove this relationship.
Here's an example of how I used STR Match Finder to predict that a group of men tracing descent to the Azores were the first J-Z2432 (otherwise South and West Asian) verified by NGS testing to be found in Europe.
An Azorean J-Z8326 is the First J-Z2432 Found in Europe – His Ancestors Migrated From India