The closest matches found by STR Match Finder are not necessarily actually the men who are most closely related to you or another man whose STRs you queried on.
Other tutorials explain how to look for shared, rare alleles, indicated in shades of red, to identify men more likely to share more recent common descent.
This tutorial focuses on how to remove men from the list who are noise - they aren't actually related to you but show up high on your matches and may even share some rare alleles simply due to convergence, meaning a sequence of random events accumulated by their ancestors coincidentally converged to your haplotype. Haplotype means the STR allele signature of your haplogroup's MRCA, the Most Recent Common Ancestor.
Eliminate based on haplotype of your potential match
This one is the most straightforward. By inspection of the haplogroup field you can see that some men are positive for a certain haplogroup.
Understanding the part-whole relationship (who remembers WordMasters Challenge?) implied by haplogroups, sometimes a man being positive for one branch implies he must be negative for another, if the branches are in different parts of the tree rather than one being above or below the other.
Above and below in this sense refer to relative position on the tree if the top is the ancestor with descendants branching out below.
So in my haplogroup, J2-M172, we know that J2-M205 and J2-L283 are different child branches of J2-M172. If I am sure that the guy I am querying on should be in one of these two child branches, I should ignore any man of the other haplogroup.
Note that people who only tested STRs at FTDNA are only assigned their most general major haplogroup in their system even if it is obvious based on exact matches to men of more specific lineages. So keep in mind that such men could be positive for a more specific branch below the indicated haplogroup.
Check the matches of your potential match
A potential match should be excluded to you if his closest, reliable STR matches are confirmed in a different branch as explained above.
So you can query on that match's id to assume (if possible) what haplogroup he is likely to be based on his closest, reliable matches.
If you cannot make any assumption as to his haplogroup based on his matches then you cannot yet exclude him.
Your relatives should match your haplogroup's haplotype
The STR alleles that the MRCA of a haplogroup had is referred to as the haplotype.
Living descendants of the MRCA are not all guaranteed to retain this haplotype. Random mutations accumulated by a man's ancestors could obscure the STR alleles originally passed down by their common ancestor.
In one example, I predicted that a man from Denmark could be positive for a branch of J2b-Z8429 consisting completely of Tatars and Mokshas from the Volga region of Russia. I made the prediction on the basis of a few shared rare alleles.
It was not a 'smoking gun' rare STR allele signature that he shared but I considered it worth the small cost to test the SNP for the potential to make a breakthrough into this geographically outlying lineage of J2b-Z1043.
The Danish man turned out negative. Later I realized that he lacked a key STR allele that all men descending from J2b-Z8429 have, DYS485 15 to 17.
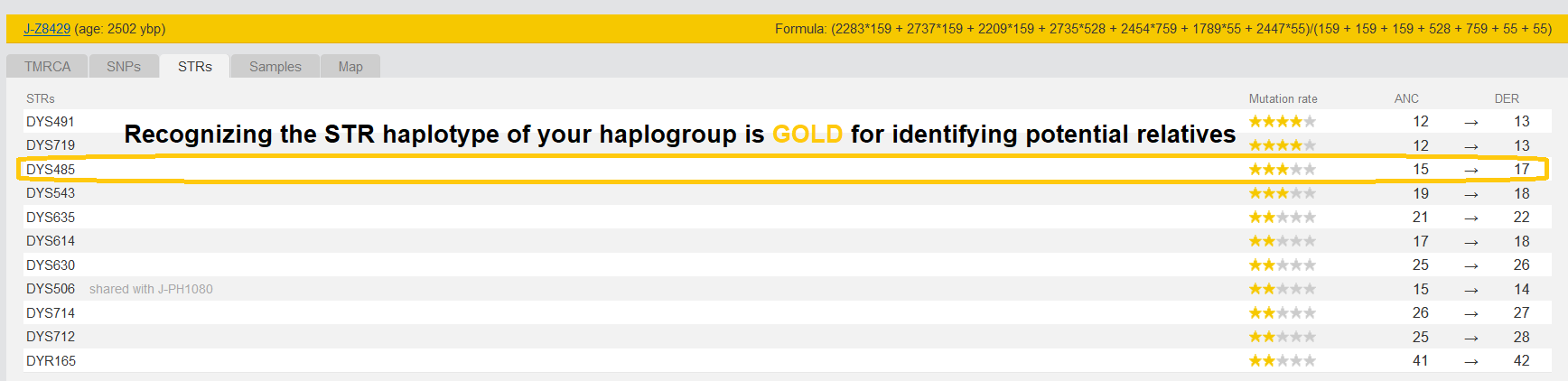
DYS485 is a relatively stable allele and the mutation that all men descending from the J2b-Z8429 MRCA have is off by two from what other men have. So while we see in some men in this lineage that the 17 mutated one more step in the last 2000+ years after the MRCA lived, none of the men in this lineage have the original allele of 15.
Fool me once, shame on me. Fool me twice? You can't fool me twice.
Advanced - Excluding because the shared rare allele was too recently acquired
A useful but more advanced technique is a variation of the "Checking the Matches" of your potential match described above in the event that the haplogroup of those men is not known.
In this case we use a different form of the process of elimination.
We can eliminate a potential match if his shared rare allele with you must have been more recently acquired than from his potential MRCA with you.
How can we recognize this?
- His closest STR matches are significantly closer than his match to you
- He shares more rare, reliable alleles with them than he does to you
- The rare allele you share with him is not from the haplotype of his MRCA
In this case, we can safely conclude that your sharing a rare allele(s) with this man is due to convergence that happened after the MRCA of this man's closer matches lived. His match to you can be ignored.