Note: HRAS is now in a state of flux as I make adjustments following the initial release. Once it becomes more stable I may cut a release version which will remain stable until you see the version number update with the next release.
One of the steps in the subclade centroid calculation involves computing a weighted average position of all immediate children nodes. Factors affecting the weighting involve the coalescence age of each child node and whether or not the sample is ancient.
HRAS computes the centroid as the midpoint of this weighted average and the “best node”, i.e. the child node whose position minimizes the total weighted distance to all other nodes.
Using the midpoint is advantageous to using the “best node” because it adds some separation on the map between subsequent subclades, allowing the subclades along the path to be more easily be visually identified, and it may more accurately reflect the true origin because of the centering effect.
However, because a very distant outlier can still have a relatively strong pulling effect, I decided to add a simplistic rule to help minimize the impact of outliers in this averaging step.
The new code can be found in this git commit – https://github.com/hprovyn/hras/commit/82c27d0e90ba02df35a73f9876f3d928033bd2f3
I define the Raw Centroid as the weighted average of latitudes and longitudes of nodes before any outlier penalty is applied.
Outlier Penalty Weight = (Nodes’ Average Distance to Raw Centroid / Sample’s Distance to Raw Centroid) ^ 2
So according to the new logic, any sample whose distance to the Raw Centroid exceeds the average will have a penalty weight applied, proportional to the square of the ratio of these distances.
In practice it has the following effect for a group of N points which have the same location and with an additional outlying point.
| N | % Raw Centroid skewed toward Outlier | % Outlier-Corrected Centroid skewed toward Outlier | % Reduction |
| 2 | 33% | 18.2% | 45.4% |
| 3 | 25% | 7.7% | 69.2% |
| 4 | 20% | 3.8% | 80.8% |
| 5 | 16.6% | 2.2% | 87% |
Note that whatever centroid above would be computed, HRAS takes the midpoint of this position with the “best node” as its “finalized” centroid which is shown on the map.
Another case to consider is how it effects a group of N points which are equidistant around a center and with an outlier that is M times further away from the center than the N points are.
| N | M | % Raw Centroid skewed toward Outlier | % Outlier-Corrected Centroid skewed toward Outlier | % skew Reduction |
| 2 | 3 | 33% | 24.4% | 26.6% |
| 2 | 5 | 33% | 20.7% | 38% |
| 2 | 10 | 33% | 18.8% | 43.5% |
| 2 | 30 | 33% | 18.3% | 45.2% |
| 3 | 3 | 25% | 16.8% | 33% |
| 3 | 5 | 25% | 10.6% | 57.4% |
| 3 | 10 | 25% | 8% | 67.9% |
| 3 | 30 | 25% | 7.7% | 69.1% |
| 4 | 3 | 20% | 10% | 50.3% |
| 4 | 5 | 20% | 5.1% | 74.6% |
| 4 | 10 | 20% | 4.1% | 79.5% |
| 4 | 30 | 20% | 3.9% | 80.1% |
For the math geeks, you may notice that the first table’s values outlier-corrected values are the limit as M goes to infinity in the second table.
This type of penalty function is good because it is applied gradually, i.e. the function is continuous. So slightly different input data does not result in a drastically different output. On the other hand, logic that would completely ignore a sample given certain criteria is not continuous.
HRAS Parameters Changed to Increase Weight of Basal and Ancient Samples for Centroid Computation
In a separate change, but committed at the same time, I have changed the parameters to increase the weight of basal and ancient samples in the computation of centroids.
I plan to eventually make these parameters configurable. For now I have tweaked them to values that should, I hope, more accurately compute the true origin of a haplogroup.
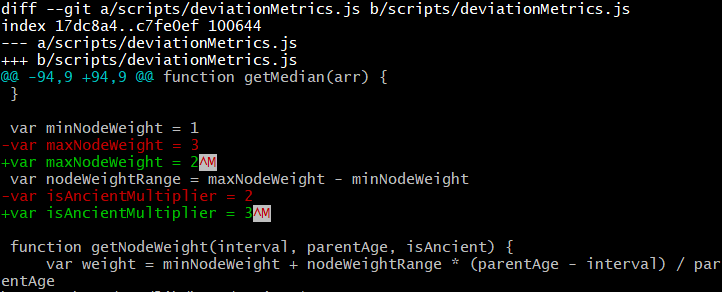
maxNodeWeight is the maximum weight that a child node can have, as a function of its coalescence age (interval variable in code below).
Previously I had this set as 3. This means that for a subclade with a 3000 year ago TMRCA, any child clade with 1500 year ago TMRCA had the weight of 1 + 2 * 1500 / 3000 = 2. A child clade with 500 year ago TMRCA had weight of 1 + 2 * 500 / 3000 = 1.33. A basal sample had weight of 1.
function getNodeWeight(interval, parentAge, isAncient) {
var weight = minNodeWeight + nodeWeightRange * (parentAge - interval) / parentAge
if (isAncient) {
return weight * isAncientMultiplier
} else {
return weight
}
}Why is the basal sample weighted less than a branch?
A basal sample is logically equivalent to a subclade with TMRCA approaching zero. So according to that formula, it gets the minimum weight.
The reason this makes sense is because the longer the coalescence age, the greater the likelihood that any given lineage may have migrated further away from the ancestral origin, assuming complete randomness.
However, on inspection, I decided that a 1/3 penalty for basal samples relative to a child subclade that coalesced immediately thereafter (i.e. same TMRCA as parent), was too much.
I believe that changing the maxNodeWeight from 3 to 2 preserves the desired effect, according to the theory stated above, and allows basal samples to more meaningfully contribute to centroid computation.
I also changed the isAncientMultiplier variable from 2 to 3 to further increase the weight of ancient samples.
The new code can be found in this git commit – https://github.com/hprovyn/hras/commit/5f54eebbb8780d09a3617363d0de639c6d9e8a81
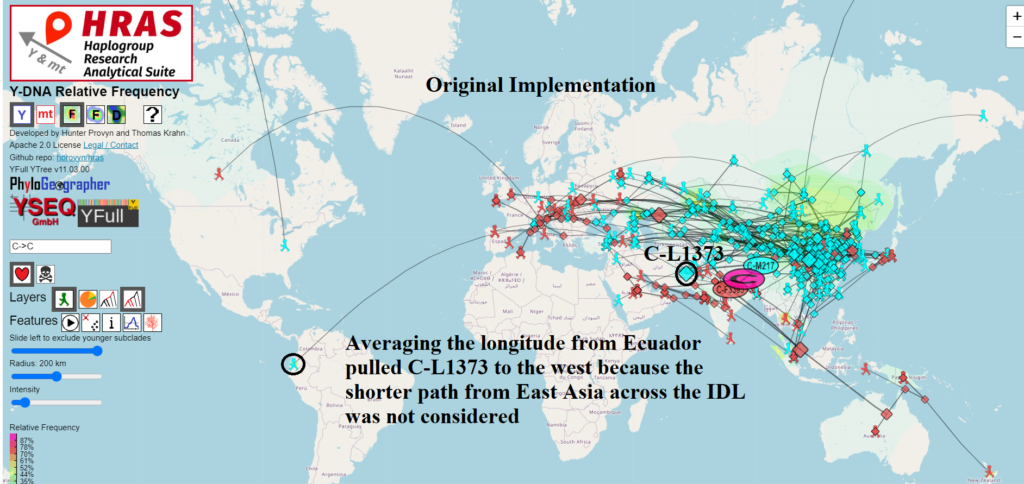
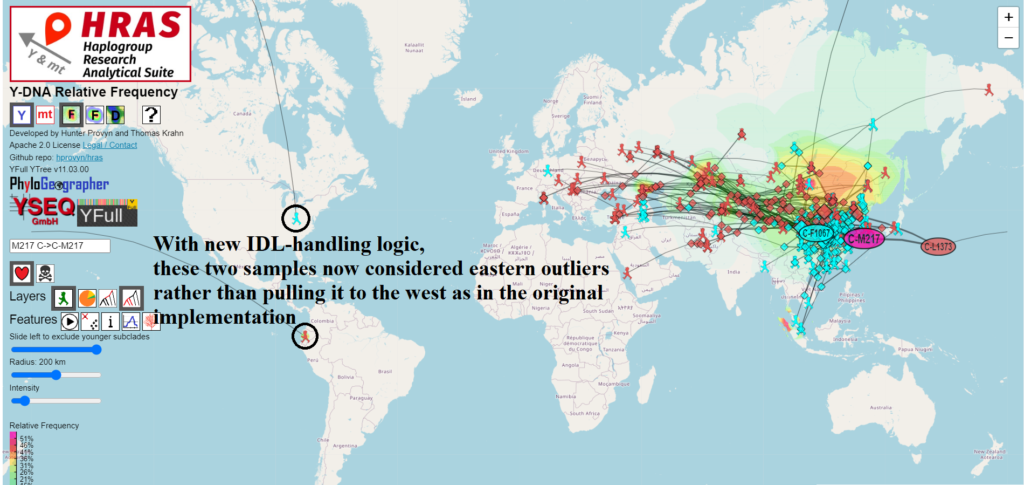
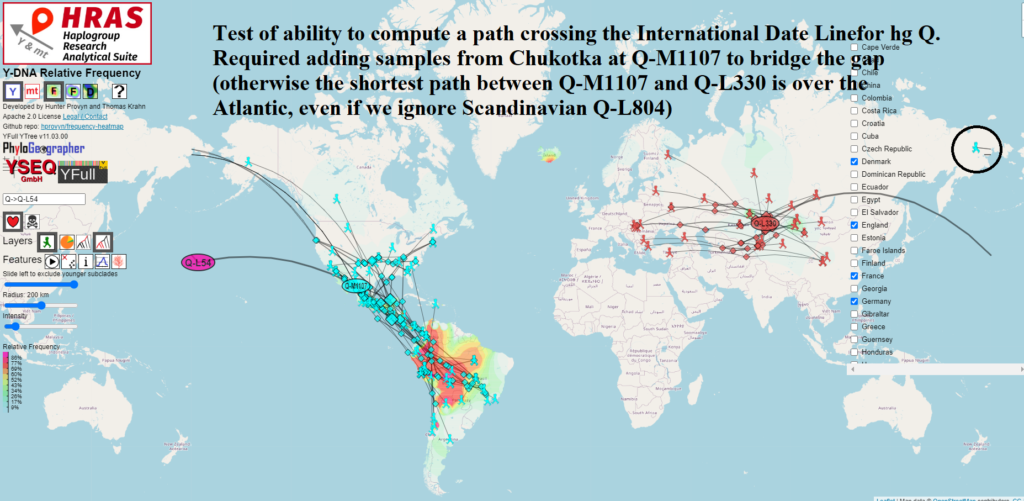
Migration Paths Can Now Cross International Date Line
For most mapping / routing applications that cover short distances, we treat the Earth as if it were flat and use Euclidean geometry functions to find the shortest distance between two points.
However the Earth is not flat and it wraps around itself if you go west from -180 degrees longitude or if you go east from 180 degrees longitude.
This place of discontinuity is called the International Date Line and mapping / routing software requires special handling to deal with crossing it.
In order to use the arithmetic mean to correctly compute the centroid of a set of points on the Earth, it is necessary to first determine which offset to use to transform the points into a 360 degree slice of longitude which is safe for averaging.
I wrote a light-weight method employing binning that achieves this and does not noticeably degrade performance of the on-the-fly calculations done in the browser.
The new code can be found in this git commit – https://github.com/hprovyn/hras/commit/d8f34b84def0136178a3ca799e63b15a1e78145e